Demo
[demo presentation] [demo recording] [report]
The SageCite demonstrator shows how the DataCite service can be used for registering DOIs for data that are generated from the building of disease network models. Workflows shared by researchers at Sage Bionetworks were captured in Taverna. The demonstrator is shown in action in this presentation and in the recording.
Two workflows were developed in the SageCite project that replicated data pipelines used at Sage Bionetworks. These pipelines are used in the metaGEO project which curates and processes microarray data from the GEO database for use in meta-analyses of gene expression from multiple microarray experiments. A plugin was developed which enables the data generated from workflows to be registered with DOIs using the DataCite service. The two Sage BIonetworks workflows are compute-intensive and take several minutes to run to completion. The DataCite plugin is demonstrated in the presentation and recording using a simpler workflow which involves retrieving a protein sequence from the NCBI database Fasta format.
The recording shows these steps:
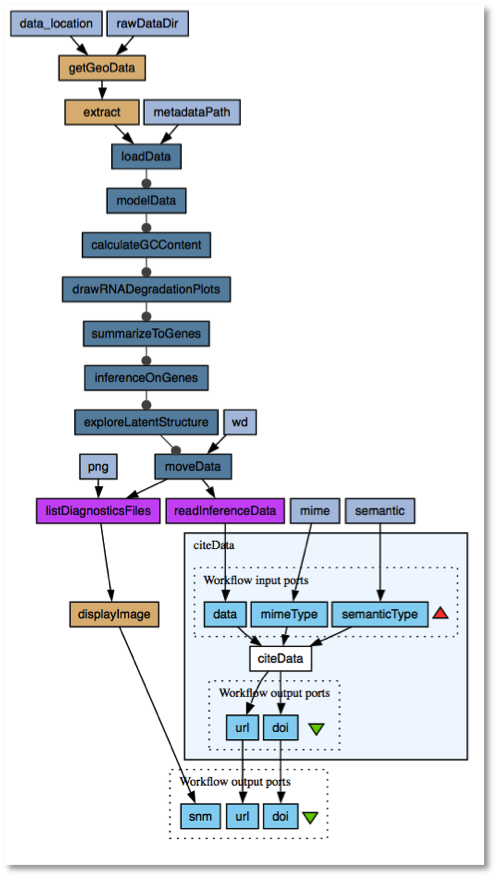
To register a workflow output with a DOI, the workflow builder selects the citeDataactivity from the service palette and adds it to the workflow diagram. The data to be registered is then fed to its data workflow input port. Additional information about the workflow can also be provided, such as its mime and semantic type. Once the workflow is built, it can be run. When the workflow is executed, the retrieved protein sequence is allocated a DOI which is used to register it using the DataCite service. The DataCite metadata store then shows the sequence registered with a DOI. DOIs have a latency it can take up to 24 hours until a DOI update is globally known; however usually after about 5 minutes, the DOI is resolvable and clickable, and resolves to the SageCite Google demo data repository web site.
A report on this development now follows.
SageCite report on the development of the workflow demonstrator by WP2
by Peter Li
Introduction
Sage Bionetworks is an organization that develops statistical models of diseases. These models represent the relationships between biological entities such as genes and proteins, and can be used to predict the behaviour and causal factors of diseases.
The work undertaken by the SageCite project required a format that was able to record and document each step of the disease model building process. This can be achieved using Taverna workflows which can explicitly record each individual step of informatics data processing pipelines used for developing statistical models. Furthermore, such workflows are executable using the Taverna workbench allowing for the results from pipelines to be reproduced.
Attempts were made to replicate the process of building disease models as Taverna workflows using the information available from the Materials and Methods section of papers published by scientists in Sage Bionetworks such as Yang et al. (2009) Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nature Genetics 41: 415-423. However, it quickly became apparent that the complexity of the model building was vastly underestimated. In addition, it was found that not enough information was present in papers or the software required was not available to replicate the modelling process. Sometimes, information was available but you were required to manually pursue it in the scientific literature due to the citation of methods previously published in past scientific publications.
SageCite workflows
The above issues led to a SageCite project visit to Sage Bionetworks in Seattle in order to understand their model development process. Following a presentation on the SageCite project, meetings were undertaken with a number of personnel and scientists including Mike Kellen, their compute platform project leader, who provided an overview of the model building process from a technology point of view.
In addition, a meeting was arranged with Brig Mecham, post-doctoral scientist at Sage Bionetworks, who agreed to provide the software tools, data and scientific knowledge to implement the data pipeline processes he is working on at Sage Bionetworks as Taverna workflows. The workflows integrated microarray data from different experiments from the Gene Expression Omnibus database .
This data pipeline developed by Brig Mecham is part of his MetaGEO project which is providing integrated gene expression datasets for meta-analyses to enable gene signature models to be determined for identification of human diseases. Whilst similar work has been done in the past, the difference of MetaGEO is that all the tools and processes are available as open source. Thus, the workflows developed as part of the SageCite project are an integral part of MetaGEO since they document exactly how the integrated MetaGEO data sets are produced. Furthermore, these workflows have been loaded into myExperiment, an online repository for sharing workflows where they are shared for the benefit of the scientific community.
Whilst it is surprising, it is also generally accepted that the data analyses cannot be reproduced based solely on information from papers (Ioannidis et al., 2009, Repeatability of published microarray gene expression analyses. Nature Genetics 41: 149-155). However, this is important if you want to re-use procedures reported in publications and to test whether you are using them correctly. Taverna workflows provide one way to document such processes in an explicit manner which can shared within the scientific community in an open science manner. It should be noted that most informatics data processes are complex and so close collaboration with the scientists responsible for their creation are required to develop such workflows.
Data citation and workflows
The availability of Taverna workflows implementing the informatics processes of Sage Bionetworks enabled the data associated with the building of disease models to be made identifiable and therefore citable. The partnership with the British Library in the SageCite project led to the use and testing of DataCite for assigning persistent identifiers in the form of Digital Object Identifiers (DOIs) to data produced from workflows. A plan was developed to enable the addition of steps into Sage Bionetworks workflows for citable data to be registered with a DOI using the DataCite service via its REST API. This was demonstrated by developing a prototype Taverna plugin using Java client code which allowed the wrapping of DataCite service. This plugin provides a DOI for a data item to be registered by DataCite as well as an URL for the SageCite demo repository where the workflow data can be downloaded.
A number of issues were encountered during this part of the work on the workflow demonstrator. In particular, there is the question of whether data should or should not be cited. This decision is made by the scientist and it depends on the extent of the value that has been added to data since it has been transformed from its initial state by the workflow. Another issue is the cost of maintaining DOIs. A scientist may want to make a large of set of data items identifiable for citation purposes but this need has to be carefully balanced with the resources required for the maintenance of DOIs.
Future work
A number of opportunities exist for further work initiated by the SageCite project. The SageCite project has shown that bioinformatics workflows and associated data can be made identifiable using a myExperiment identifier and DOIs, respectively. People are associated with these components such as the builder of the workflow and the scientists who developed the data processing pipeline implemented by the workflow. These people need to be made citable for a number of purposes such as for attribution. This could be achieved by integrating Taverna with technology being developed by the ORCID project which is a community effort to establish an open, independent registry in a way that removes name ambiguity and problems with accreditation in the scholarly research ecosystem.
Perhaps the most exciting follow-on work to the SageCite project is the use of its workflows in the Systems Science for Health (SSFH) project at the University of Birmingham. The MetaGEO workflows implemented by SageCite are generic in that they can be used for integrating microarray data from a variety of laboratory experiments. Due to the generic nature of the workflows, they are currently being applied to the study of blood cancers and human diseases of inflammation such as rheumatoid arthritis which are under investigation by SSFH. It is hoped that this collaboration with Sage Bionetworks can be maintained by this line of work.